Matheus G Mussi
University of Alberta (Edmonton)
INTRODUCTION
People experiencing neurological disorder can have severe limitations in their functional abilities [1]. They usually rely on assistive technology to perform daily activities [2], but sometimes they are difficult to control. Brain-computer interfaces (BCI) are a potential assistive technology solution. This technology is feasible for people with complex physical needs because in many cases, the disorders do not affect the cognition extensively [3–7]. Traditional BCI methods, nevertheless, yield insufficient performance to be used in real-time applications and are hard to operate independently, which are important criteria for end-users [8,9]. Individuals experience fatigue when low accuracy rates require them to repeat entries and correct selections. Traditional BCI rely on a single input signal (e.g. EEG), single source of stimulus (e.g. auditory, visual, tactile, etc) or a single brain signal paradigm (pattern). This causes the system to have low information transfer rate, an inflexible human-interface and less information to improve its performance [10]. Hybrid brain-computer interfaces aim to improve on performance and speed through multi-modal signal inputs, combining different brain-signals, BCI paradigms or other external device input [11].
There are many hybrid brain-computer interface (hBCI) systems that use various combinations of brain signals and physiological signals or other devices. Nonetheless, very few papers explain the details of how to 'assemble' an hBCI. This paper explains the development of software capable of displaying simultaneous stimuli of SSVEP and P300 which will be used in future research with children with disabilities.
ARCHITECTURE
Interface
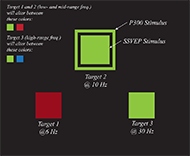
For our system's display, there will be three squares that have coloured centre areas that flash at different frequencies for the SSVEP stimuli, and an outline edge that will appear around the squares one at a time in a pseudo-random order for the P300 stimulus, as shown in Fig. 1.
For the SSVEP, squares will create the flashing effect by interpolating between colors, as proposed by [12]. The chosen frequencies are 30, 10 and 6 Hz, to avoid seizure-inducing ranges. When the classification is concluded, the selected square's centre area will briefly turn white in colour to indicate the classifier chose that square.
Program
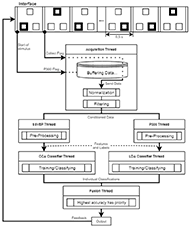
For the online operation of the system, threads are needed to avoid delays in the execution of processes that need to be concurrent. The architecture of the program is based on a main section that initializes other threads and variables, the experiment thread, which displays the squares on the screen using Psychopy, the acquisition thread, the feature extraction threads (SSVEP and P300 specific) and the classification threads. The overall architecture scheme can be seen in Fig. 2.
To synchronize the functioning of the different paradigms, the program is based on threading event and standardized intervals. Every half second, a new P300 stimulus is presented and the buffered data is pre-processed and sent to the classifier for training or validation. Events are sent to the active threads and their function is synchronized, avoiding data loss. An important note on threading events is that they have a built-in wait function, which avoids unnecessary loops and protects Python's synchronization primitives.
To transfer data from one thread to the other, threading queues are used. They are needed because Global variables are not supported by threads. Queues have a first-in-first-out method and they are emptied every time the get method is used.
DEVELOPMENT
The developed system is based on Python3®. The libraries used are freely available online and the software should be compatible with many BCI headsets, depending primarily on the BrainFlow library. This system was tested with the OpenBCI Cython board. The system runs in a Windows 10 operational system, but most of the installations should also work with Linux. Mac users might experience some compatibility issues with some headsets (especially due to the deprecation of Future Technology Devices International (FTDI) chip drivers, which hinders the use of the OpenBCI board).
Setting up the experiment on a new computer
This sub-section explains the installation of all the necessary resources to create a hBCI system with visual stimulation. The steps below are meant to be done in sequence.
Requirements
The basic requirements for the system are: 1) Python 3.6.6, 2) Anaconda3 and 3) Psychopy3. Python 3.6.6 is currently the most up-to-date version that supports all the required packages. In addition to Python, Anaconda was used to maintain all the specific libraries for this system separated from other libraries in the operational system. Psychopy3 is a package used to manipulate screen elements to create stimuli for the users.
Creating an environment with Anaconda with Psychopy3
Environments are directories containing specific packages which do not affect other environments. This gives the ability of installing packages and package versions that are project-specific [13]. For this project, the environment is based on an environment containing Pychopy3. This method avoids compatibility issues.
Firstly, the Anaconda3 executable [14] and the Psychopy3 environment file (psychopy-env.yml) [15] need to be downloaded. Anaconda needs then to be installed and the Psychopy file saved in an accessible folder. Once anaconda3 is installed, through the Anaconda Powershell, navigate to the folder containing the Psychopy file and enter conda env create –n psychopy –f psychopy-env.yml to create the environment named "psychopy". To activate the environment, in the Powershell, enter conda activate psychopy.
Once the environment is running, some packages are needed (some packages are OpenBCI Cython-specific). Most of them need to be installed via conda install –c command, but some are installed through pip install. The packages are cython, pyserial, matplotlib, piglet (v.1.4.10), numpy, pygtgraph, scipy, pandas, pylsl, and Pillow.
The specific Pyglet version needs to be 1.4.10 instead of newer versions because some later versions yield the error "WMF Failed to initialize threading: err.strerror" when using threads. Depending on the application, some dll files can be missing and also generate errors. If the errors "The program can't start because VCRUNTIME140D.dll is missing from your computer" or "The program can't start because ucrtbased.dll is missing from your computer" appear, the dll's need to be downloaded and pasted in the C:/Windows/System32 folder.
Installing BrainFlow
BrainFlow is a library to parse and obtain biosensory data from devices. This library allows an easy connectivity between electroencephalography (EEG) headsets and the computer/program. The strengths of this library over others are its easy implementation, its board versatility (e.g. a few lines of code can easily adapt the program to receive signals from OpenBCI Cython or g.tec Unicorn) and its vivid community, which gives vast support through Slack. To install BrainFlow in the environment, enter python –m pip install brainflow.
Installing Scikit-learn
Scikit-learn is a library for predictive and data analysis. It provides simplicity and a vast range of tools to classify and analyse data. The classifiers in this library will be implemented for the system's decision making. It can also be installed in the environment with conda install scikit-learn.
Remove excessive signal latency
In some boards, the default latency can cause the signal to be read incorrectly or in chunks. To avoid these, the latency needs to be decreased. This can be done by going to the Device Manager (Right click on My Computer/This PC, then Properties and select Device Manager). Then, the port setting needs to be adjusted: expand the Ports (COM & LPT) list, right click the serial port corresponding to the device (e.g. COM3) and select Properties. In Properties, go to the Port Settings tab, then Advanced, and then change Latency Timer (msec) to 1.
Working with BrainFlow
BrainFlow gives the developer the ability to connect and start streaming from within the code. The minimum sequence to connect to a board is the following:
import brainflow
from brainflow.board_shim import BardShim, BrainFlowInputParams, BoardIds
from brainflow.data_filter import DataFilter, FilterTypes, AggOperation
BoardShim.enable_dev_board_logger() params = BrainFlowInputParams()
params.serialport = 'COM3'
board_id = BoardIds.CYTON_BOARD.value #variable for specific board
board = BoardShim(board_id, params) #creates callable object
board.prepare_session() #finds and connects to board
board.start_stream() #starts streaming
data = board.get_board_data() #collects data
Each board needs a specific set of parameters. To see what parameters are needed, refer to [16]. For the board_id variable, the board in use should be called (e.g. if using the g.tec Unicorn, use board_id = BoardIds.UNICORN_BOARD.value). The data streamed from the board comes in a single array which contains timestamps and sensory data.
FUTURE RESEARCH
This hBCI is currently undergoing testing and the next steps are 1) tests with adults with and without disabilities, using a User Centred Design approach to improve the system and 2) experiments with children with disabilities to see its efficacy in clinical applications.
REFERENCES
- Bauer G, Gerstenbrand F, Rumpl E. Varieties of the locked-in syndrome. J Neurol. 1979 Aug 1;221(2):77–91.
- Cook AM, Polgar JM. Assistive technologies : principles and practice [Internet]. 2014. Available from: https://search.ebscohost.com/login.aspx?direct=true&scope=site&db=nlebk&db=nlabk&AN=1151635
- Stadskleiv K. Cognitive functioning in children with cerebral palsy. Dev Med Child Neurol. 2020;62(3):283–9.
- Mahon S, Faulkner J, Barker-Collo S, Krishnamurthi R, Jones K, Feigin V. Slowed Information Processing Speed at Four Years Poststroke: Evidence and Predictors from a Population-Based Follow-up Study. J Stroke Cerebrovasc Dis. 2020 Feb 1;29(2):104513.
- Cioni G, Guzzetta A, D'Acunto G. Cerebral Plasticity and Functional Reorganization in Children with Congenital Brain Lesions. In: Buonocore G, Bracci R, Weindling M, editors. Neonatology: A Practical Approach to Neonatal Diseases [Internet]. Milano: Springer Milan; 2012 [cited 2020 Sep 9]. p. 145–9. Available from: https://doi.org/10.1007/978-88-470-1405-3_21
- Keller J, Böhm S, Aho-Özhan HEA, Loose M, Gorges M, Kassubek J, et al. Functional reorganization during cognitive function tasks in patients with amyotrophic lateral sclerosis. Brain Imaging Behav. 2018 Jun;12(3):771–84.
- Sabbah P, Schonen S de, Leveque C, Gay S, Pfefer F, Nioche C, et al. Sensorimotor Cortical Activity in Patients with Complete Spinal Cord Injury: A Functional Magnetic Resonance Imaging Study. J Neurotrauma. 2002 Jan;19(1):53–60.
- Huggins JE, Wren PA, Kirsten L. Gruis. What would brain-computer interface users want? Opinions and priorities of potential users with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011 Sep 1;12(5):318–24.
- Collinger JL, Boninger ML, Bruns TM, Curley K, Wang W, Weber DJ. Functional priorities, assistive technology, and brain-computer interfaces after spinal cord injury. J Rehabil Res Dev. 2013;50(2):145.
- Zina Li, Shuqing Zhang, Jiahui Pan. Advances in Hybrid Brain-Computer Interfaces: Principles, Design, and Applications. Comput Intell Neurosci [Internet]. 2019 [cited 2020 Feb 23]; Available from: https://www.hindawi.com/journals/cin/2019/3807670/
- Wolpaw J, Wolpaw EW. Brain–Computer Interfaces: Principles and Practice. Oxford University Press; 2012. 424 p.
- Floriano A, Diez PF, Freire Bastos-Filho T. Evaluating the Influence of Chromatic and Luminance Stimuli on SSVEPs from Behind-the-Ears and Occipital Areas. Sensors [Internet]. 2018 Feb 17 [cited 2020 Oct 8];18(2). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5855130/
- Conda environments [Internet]. [cited 2021 Feb 13]. Available from: https://docs.conda.io/projects/conda/en/latest/user-guide/concepts/environments.html
- Anaconda - Individual Edition [Internet]. Anaconda. [cited 2021 Feb 13]. Available from: https://www.anaconda.com/products/individual
- Installation - Anaconda and Miniconda [Internet]. Psychopy3. [cited 2021 Feb 13]. Available from: https://www.psychopy.org/download.html#anaconda-and-miniconda
- Supported Boards - BrainFlow documentation [Internet]. [cited 2021 Feb 13]. Available from: https://brainflow.readthedocs.io/en/stable/SupportedBoards.html